Format for reports of the results of simulation experiments described with SED-ML
Overview
The BioSimulators/BioSimulations format for simulation results outlines how SED-ML reports (sedml:report) and plots (sedml:plot2D, sedml:plot3D) should be encoded into Hierarchical Data Format (HDF) 5. These conventions are capable of capturing reports and plots with multiple dimensions, and with data sets that have different shapes and data types, and repeated labels.
Specifications
Data for reports and plots of simulation results should be saved in HDF5 according to the following conventions:
-
Paths of reports and plots: Within the HDF5 file, each report/plot should be saved to a path equal to the combination of (a) the relative location of the parent SED-ML document within the parent COMBINE/OMEX archive and (b) the
idof the report/plot. For example, a report with idtime_course_resultsin a SED-ML file located at./path/to/experiment.sedmlshould be saved to the pathpath/to/experiment.sedml/time_course_resultsin the HDF5 file. -
Data set shapes: For SED-ML reports, the rows of each HDF5 dataset should correspond to the SED-ML data sets (
sedml:dataSet) specified in the SED-ML definition of the report (e.g., time symbol, specific model variables). For SED-ML plots, the rows of each HDF5 dataset should correspond to the SED-ML data generators (sedml:dataGenerator) specified in the SED-ML definition of the plot (e.g., time symbol, specific model variables). -
Elemental tasks (
sedml:task):- Steady-state simulations (
sedml:steadyState): The rows of HDF5 data sets should be scalars. - One step simulations (
sedml:oneStep): The rows of HDF5 data sets should be tuples of the start and end points of the simulation. - Time course simulations (
sedml:uniformTimeCourse): The rows of HDF5 data sets should be a vector with length equal to the number of steps of the time course plus one. - Simulations of spatial models: The rows of HDF5 data sets should be matrices whose dimensions represent space and time.
- Steady-state simulations (
-
Repeated tasks (
sedml:repeatedTask): The first dimension of each row should represent the iterations of the tasks that produced its values. The second dimension of each data set should represent the individual sub-tasks (sedml:subTask) of the task. The results of sub-tasks should be ordered in the same order -- the order of their attributes -- that the sub-tasks were executed. If repeated tasks are nested within repeated tasks, the next dimensions should alternate between representing the iterations and sub-tasks of the nested repeated tasks. The final dimensions of each row should be encoded as above forsedml:task. For example, non-spatial time course simulations should have a single additional dimension of length equal to the number of steps of the time course plus one.
Note
If the rows of an HDF5 data set have different shapes, the data sets should be reshaped into a consistent shape by right-padding their values with NaN.
-
Metadata for reports: The following metadata should be encoded into attributes of the corresponding HDF5 dataset.
- Type of the output: The type of the output (
Report,Plot2D,Plot3D) should be encoded into the key_type. - Complete id of the output: The complete id of the output (combination of the location of the parent SED-ML file of the output (
omex-manifest:content/@location) within its parent COMBINE/OMEX archive and the SED-ML id of the output (sedml:output/@sedml:id)) should be encoded into the keyuri. - Id of the output: The SED-ML id of the output (
sedml:output/@sedml:id) should be encoded into the keysedmlId. - Name of the output: The name of the output (
sedml:output/@sedml:name) should be encoded into the keysedmlName. - Ids of rows (SED-ML data sets or data generators): For reports, the ids of the data sets should be encoded into the key
sedmlDataSetIds. The value of this key should be an array of the ids of the data sets, in the order in which the data sets were defined in their parent SED-ML document. For plots, the ids of the data generators should be encoded into the keysedmlDataSetIds. The value of this key should be an array of the ids of the data generators, in the order in which the data generators were defined in their parent SED-ML document. - Names of row (SED-ML data sets or data generators): For reports, the names of the data sets should be encoded into the key
sedmlDataSetNames. For plots, the names of the data generators should be encoded into the keysedmlDataSetNames. The value of this key should be an array of the ids of the data sets, in the order in which the data sets were defined in their parent SED-ML document. - Labels of rows (SED-ML data sets or data generators): For reports, the labels of the data sets should be encoded into the key
sedmlDataSetLabels. For plots, the id of the data generators should be encoded into the keysedmlDataSetLabels. The value of this key should be an array of the labels of the data sets, in the order in which the data sets were defined in their parent SED-ML document. - Data types of SED-ML data sets/generators: The data types of the data sets (reports) or data generators (plots) should be encoded into the key
sedmlDataSetDataTypes. The value of this key should be an array of the data types of the data sets/generators, in the order in which the data sets/generators were defined in their parent SED-ML document. The data type of each data set should either be described using a NumPydtype(e.g.,int64) to indicate a data set whose value is non-null or__None__to indicate a data set whose value isnull. - Shapes of SED-ML data sets/generators: The shapes of the data sets (reports) or data generators (plots) should be encoded into the key
sedmlDataSetShapes. The value of this key should be an array of comma-separated lists of the shapes of the data sets/generators. The shapes of the data sets/generators should be listed in the order in which the data sets/generators were defined in their parent SED-ML document.
- Type of the output: The type of the output (
-
Metadata for SED-ML files: The following metadata should be encoded into attributes of the parent groups of HDF5 datasets which represent SED-ML files and their parent directories within their parent COMBINE/OMEX archives.
- Complete id of the COMBINE/OMEX archive location: The location of each SED-ML file and the location of each parent directory of each SED-ML file with their parent COMBINE/OMEX archive (
omex-manifest:content/@location) should be encoded into the keysuriandcombineArchiveLocation.
- Complete id of the COMBINE/OMEX archive location: The location of each SED-ML file and the location of each parent directory of each SED-ML file with their parent COMBINE/OMEX archive (
Example HDF5 report files
Several example reports are available here.
Below is a graphical illustration of the organization of an HDF5 file for a SED-ML report with id report-1 defined in a SED-ML file located at experiment-1/batch-1/simulation-1.sedml within a COMBINE/OMEX archive.
Path of the HDF5 dataset for the SED-ML report
experiment-1/batch-1/simulation-1.sedml/report-1
HDF5 dataset for the SED-ML report
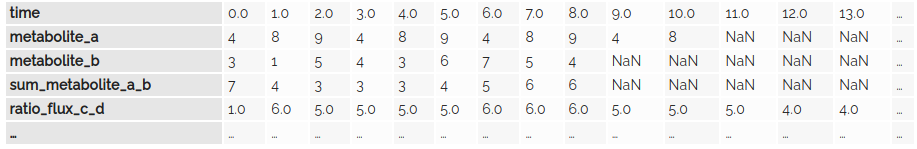
Attributes of the HDF5 dataset for the SED-ML report
_type:Reporturi:experiment-1/batch-1/simulation-1.sedml/report-1sedmlId:report-1sedmlName:Report 1sedmlDataSetIds:time,metabolite_a,metabolite_b,sum_metabolite_a_b,ratio_flux_c_dsedmlDataSetLabels:Time,Metabolite A,Metabolite B,Sum of metabolites A and B,Flux ratio of reactions C and DsedmlDataSetDataTypes:float64,int64,int64,int64,float64sedmlDataSetShapes:14,9,11,9,14
Attributes of the HDF5 groups for the SED-ML file and its parent subdirectories
experiment-1HDF5 group for the grandparent directory of the SED-ML fileuri:experiment-1combineArchiveLocation:experiment-1experiment-1/batch-1HDF5 group for the parent directory of the SED-ML fileuri:experiment-1/batch-1combineArchiveLocation:experiment-1/batch-1experiment-1/batch-1/simulation-1.sedmlHDF5 group for the SED-ML fileuri:experiment-1/batch-1/simulation-1.sedmlcombineArchiveLocation:experiment-1/batch-1/simulation-1.sedml
Recommended resources for building reports of simulation results
Below are helpful tools for building reports of simulation results:
- BioSimulators utils is a Python library which provides functions for generating reports to the above specifications.
- h5py is a high-level Python library for reading and writing HDF5 files.
- HDF5 libraries for C, C++, and Java.